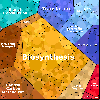
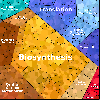
Proteomaps show the quantitative composition of proteomes.
Each protein is represented by a polygon: areas reflect protein abundance,
and functionally related proteins are arranged in common and similarly colored regions.
For further reading please see
www.proteomaps.net
user template previews
Gena Rosenberger
Struggling with low website leads for Uni Greifswald De? This short video shows how our AI can boost your traffic: https://www.youtube.com/shorts/d7kGxfGVyNg
17717750486966
Pseudomonas putida KT2440 KO (adapted by N 12072024)
BAE KT2440 upregulation list _except none
17701232562404