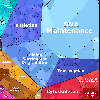
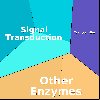
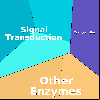
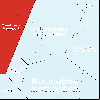
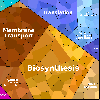
Proteomaps show the quantitative composition of proteomes.
Each protein is represented by a polygon: areas reflect protein abundance,
and functionally related proteins are arranged in common and similarly colored regions.
For further reading please see
www.proteomaps.net
user template previews
Jimmie Munger
Our AI-powered service brings precise, keyword-driven traffic from your chosen locations to your website, all at a fraction of the cost of paid ads. Connect with us today.
https://marketingaged.com/
https://marketingaged.com/
17703869775118
Monserrate Bate
Need more clicks and conversions for uni-greifswald.de? Watch this short video about our AI-powered traffic service: https://www.youtube.com/shorts/iojvp6ZtjW4
17703869750422
Wilbert Easterby
Our AI-optimized solution brings keyword and location-targeted traffic to your website, offering better value than costly paid advertising. Reach out to see results.
https://ow.ly/qTuV50XSIKn
https://ow.ly/qTuV50XSIKn
17702756487578
Pseudomonas putida KT2440 KO (adapted by N 12072024)
BAE KT2440 upregulation list _except none
17701232562404